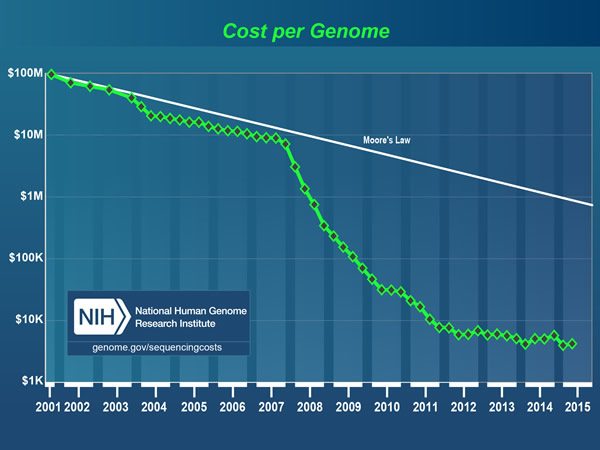
I am fortunate to have become a physician in a time of great scientific progress. Back when I was in college and medical school, the thought that we would one day be able to sequence the human genome (and now sequence hundreds of cancer genomes), to measure the expression of every gene in the genome simultaneously on a single “gene chip,” and to assess the relative abundance of every RNA transcript, coding and noncoding (such as microRNAs) simultaneously through next generation sequencing (NGS) techniques was considered, if not science fiction, so far off in the future as to be unlikely to impact medicine in my career. Yet here I am, mid-career, and all of these are a reality. The cost of rapidly sequencing a genome has plummeted. Basically, the first human genome cost nearly $3 billion to sequence, while recent developments in sequencing technology have brought that cost down to the point where the “$1,000 genome” is within sight, if not already here, as illustrated in the graph above published by the National Human Genome Research Institute. Whether the “$1,000 genome” is truly here or not, the price is down to a few thousand dollars. Compare that to the cost of, for instance, the OncoType DX 21-gene assay for estrogen receptor-positive breast cancer, which costs nearly $4,000 and is paid for by insurance because its results can spare many women from even more expensive chemotherapy.
So, ready or not, genomic medicine is here, whether we know enough or not to interpret the results in individual patients and use it to benefit them, so much so that President Obama announced a $215 million plan for research in genomic mapping and precision medicine known as the Precision Medicine Initiative. Meanwhile, the deeply flawed yet popular 21st Century Cures bill, which passed the House of Representatives, bets heavily on genomic research and precision medicine. As I mentioned when I discussed the bill, it’s not so much the genomic medicine funding that is the major flaw in the bill but rather its underlying assumption that encouraging the FDA to decrease the burden of evidence to approve new drugs and devices will magically lead to an explosion in “21st century cures,” the same old antiregulatory wine in a slightly new bottle. Be that as it may, one way or the other, the federal government is poised to spend lots of money on precision medicine.
Because I’m a cancer doctor, and, if there’s one area in medicine in which precision medicine is being hyped the hardest, it’s hard for me not to think that the sea change that is going on in medicine really hit the national consciousness four years ago. That was when Walter Isaacson’s biography of Steve Jobs revealed that after his cancer had recurred as metastatic disease in 2010. Jobs had consulted with research teams at Stanford, Johns Hopkins, and the Broad Institute to have the genome of his cancer and normal tissue sequenced, one of the first twenty people in the world to have this information. At the time (2010-2011), each genome sequence cost $100,000, which Jobs could easily afford. Scientists and oncologists looked at this information and used it to choose various targeted therapies for Jobs throughout the remainder of his life, and Jobs met with all his doctors and researchers from the three institutions working on the DNA from his cancer at the Four Seasons Hotel in Palo Alto to discuss the genetic signatures found in Jobs’ cancer and how best to target them. Jobs’ case, as we now know, was a failure. However much Jobs’ team tried to stay one step ahead of his cancer, the cancer caught up and passed whatever they could do.
That’s not to say that there haven’t been successes. For instance, in 2012 I wrote about Dr. Lukas Wartman, at the time a recently-minted oncologist who had been diagnosed with acute lymphoblastic leukemia as a medical student, was successfully treated, but relapsed five years later. He underwent an apparently successful bone marrow transplant, but recurred again. At that point, there appeared to be little that could be done. However, Dr. Timothy Ley at the Genome Institute at George Washington University decided to do something radical. He sequenced the genes of Wartman’s cancer cells and normal cells:
The researchers on the project put other work aside for weeks, running one of the university’s 26 sequencing machines and supercomputer around the clock. And they found a culprit — a normal gene that was in overdrive, churning out huge amounts of a protein that appeared to be spurring the cancer’s growth.
That was 2011 as well. Today, the sequence could have been done much more rapidly. In any case, Ley identified a gene that was overactive and could be targeted by a new drug for kidney cancer. His cancer went into remission. Wartman is now the assistant director of cancer genomics at Washington University.
The technology now, both in terms of sequencing and bioinformatics, has advanced enormously even since 2011. With it has advanced the hype. But how much is hype and how much is really hope? Let’s take a look. Also, don’t get me wrong. I do believe there is considerable promise in precision medicine. However, having personally begun my research career in the 1990s, when angiogenesis inhibitors were being touted as the cure to all cancer (and we know what happened there), I am also skeptical that the benefits can ever live up to the hype.
The origin of “precision” medicine
“Precision medicine” is now the preferred term for what used to be called “personalized medicine.” From my perspective, it is a more accurate description of what “personalized medicine” meant, given that many doctors objected to the term because they felt that every good doctor practices personalized medicine. Even so, “precision medicine” is no less a marketing term than was “personalized medicine.” If you don’t believe this, look at the hype on the White House website:
Today, most medical treatments have been designed for the “average patient.” In too many cases, this “one-size-fits-all” approach isn’t effective, as treatments can be very successful for some patients but not for others. Precision medicine is an emerging approach to promoting health and treating disease that takes into account individual differences in people’s genes, environments, and lifestyles, making it possible to design highly effective, targeted treatments for cancer and other diseases. In short, precision medicine gives clinicians new tools, knowledge, and therapies to select which treatments will work best for which patients.
If you think this sounds like what alternative medicine quacks (but I repeat myself) routinely say about “conventional medicine,” you’d be right. It’s not that precision medicine advocates don’t have a germ of a point, but they fail to put it this criticism into historical context. Medicine has always been personalized or “precision.” It’s just that in the past the only tools we had to personalize our care were things like family history, comorbid conditions, patient preferences, and aspects of the patient’s history that might impact which treatment would be most appropriate. In other words, our tools to personalize care weren’t that “precise,” making our precision far less than we as physicians might have liked. Genomics and other new sciences offer the opportunity to change that, but at the cost that too much information will paralyze decision making. Still, at its best, precision medicine offers the opportunity to “personalize” medicine in a science-based manner, rather than the “make it up as you go along” and “pull it out of my nether regions” method of so many alternative medicine practitioners. It could also offer the clinical trials tools to do it, such as NCI-MATCH. At its worst, precision medicine is companies jumping the gun and selling genomic tests direct to the consumer without having an adequate scientific basis to know what they mean or what should be done with the results.
In any case, up until 2011, the term “personalized” medicine tended to be used to describe a form of medicine not yet in existence in which the each patients’ unique genomic makeup would serve as the basis to guide therapies. Then, the National Academy of Sciences Committee issued a report, “Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease“, which advocated the term “precision medicine” and differentiated it from “personalized medicine” thusly:
“Personalized medicine” refers to the tailoring of medical treatment to the individual characteristics of each patient. It does not literally mean the creation of drugs or medical devices that are unique to a patient, but rather the ability to classify individuals into subpopulations that differ in their susceptibility to a particular disease or their response to a specific treatment. Preventive or therapeutic interventions can then be concentrated on those who will benefit, sparing expense and side effects for those who will not. (PCAST 2008) This term is now widely used, including in advertisements for commercial products, and it is sometimes misinterpreted as implying that unique treatments can be designed for each individual. For this reason, the Committee thinks that the term “Precision Medicine” is preferable to “Personalized Medicine” to convey the meaning intended in this report.
As I said, “precision medicine” is a marketing term, but it’s actually a better marketing term than “personalized medicine” because it is closer to what is really going on. That’s why I actually prefer it to “personalized medicine,” even though I wish there were a better term. Whatever it is called, however, the overarching belief that precision medicine is the future of medicine has led to what has been called an “arms race” or “gold rush” among academic medical centers to develop precision medicine initiatives, complete with banks of NGS machines, new departments of bioinformatics and genomics, and, of course, big, fancy computers to analyze the many petabytes of data produced, so much data that it’s hard to have enough media upon which to store it and we don’t know what to do with it. Genomic sequencing is producing so much data that IBM’s Watson is being used to analyze cancer genetics. It’s not for nothing that precision medicine is being likened to biology’s “moon shot“—and not always in a flattering way.
So what is the real potential of precision medicine?
Complexity intrudes
I discussed some of the criticism of precision medicine when I discussed the 21st Century Cures Act three weeks ago. I’ll try to build on that, but after a brief recap. Basically, I mentioned that I was of a mixed mind on the bill’s emphasis on precision medicine, bemoaning how now, at arguably the most exciting time in the history of biomedical research, the dearth of funding means that, although we’ve developed all these fantastically powerful tools to probe the deepest mysteries of the genome and use the information to design better treatments, scientists lack the money to do so. I even likened the situation to owning a brand new Maserati but there being no gasoline to be found to drive it, or maybe having the biggest, baddest car of all in the world of Mad Max but having to fight for precious gasoline to run it. I also noted that I thought precision medicine was overhyped (as I am noting again in this post), referencing skeptical takes on precision medicine in recent op-eds by Michael Joyner in The New York Times, Rita Rubin in JAMA declaring precision medicine to be more about politics, Cynthia Graber in The New Yorker, and Ronald Bayer and Sandro Galea in The New England Journal of Medicine. Basically, the number of conditions whose outcome can be greatly affected by targeting specific mutations is relatively small, far smaller than the impact likely would be from duller, less “sexy” interventions, such as figuring out how to get people to lose weight, exercise more, and drink and smoke less. The question is whether focusing in the genetic underpinnings of disease will provide the “most bang for the buck,” given how difficult and expensive targeted drugs are to develop.
Over the weekend, there was a great article in The Boston Globe by Sharon Begley entitled “Precision medicine, linked to DNA, still too often misses“, that gives an idea of just how difficult reaching this new world of precision medicine will be. It’s the story of a man named John Moore, who lives in Apple Valley, UT. Moore has advanced melanoma and participated in a trial of precision medicine for melanoma. His outcome shows the promise and limitations of such approaches:
Back in January, when President Obama proposed a precision medicine initiative with a goal of “matching a cancer cure to our genetic code,” John Moore could have been its poster child. His main tumors were shrinking, and his cancer seemed to have stopped spreading because of a drug matched to the cancer’s DNA, just as Obama described.
This summer, however, after a year’s reprieve, Moore, 54, feels sick every day. The cancer — advanced melanoma like former president Jimmy Carter’s — has spread to his lungs, and he talks about “dying in a couple of months.”
The return and spread of Moore’s cancer in a form that seems impervious to treatment shows that precision medicine is more complicated than portrayed by politicians and even some top health officials. Contrary to its name, precision medicine is often inexact, which means that for some patients, it will offer false hope rather than a cure.
On the other hand, in the Intermountain study, after two years, progression-free survival in the group with advanced cancer treated using precision medicine techniques was nearly twice what it was in those who underwent standard chemotherapy, 23 months versus 12 months. Moore himself reports that with a pill he had one year of improved health and quality of life before his cancer started progressing again. It’s not yet clear in this trial whether this will translate into an improvement in overall survival, the gold standard endpoint, but it’s a very promising start. It is, however, not a miraculous start.
Here’s the problem. I’ve alluded to it before. Cancer genomes are messed up. Really messed up. And, as they progress, thanks to evolution they become even more messed up, and messed up in different ways, so that the tumor cells in one part of a tumor are messed up in a different way than the tumor cells in another part of the tumor, which are messed up in a different way than the metastases. It’s called tumor heterogeneity.
Now enter the problem in determining which mutations are significant (commonly called “driver” mutations) and which are secondary or “just along for the ride” (commonly called “passenger” mutations):
But setbacks like Moore’s show that genetic profiling of tumors is, at this point, no more a cure for every cancer than angiogenesis inhibitors, which cut off a tumor’s blood supply, or other much-hyped treatments have been.
A big reason is that cancer cells are genetically unstable as they accumulate mutations. As a result, a biopsy might turn up dozens of mutations, but it is not always clear which ones are along for the ride and which are driving the cancer. Only targeting the latter can stop a tumor’s growth or spread.
Knowing which mutation is the driver and which are passenger mutations is so complicated that the Intermountain researchers established a “molecular tumor board” to help.
Composed of six outside experts in cancer genomics, the board meets by conference call to examine the list of a patient’s tumor mutations and reach a consensus about which to target with drugs. Tumor profiling typically finds up to three driver mutations for which there are known drugs, and the board reviews data on how well these drugs have worked in other patients with similar tumors.
And:
The next difficulty, Nadauld said, is that “the mutations may be different at different places in a tumor.” But oncologists are reluctant to perform multiple biopsies. The procedures can cause pain and complications such as infection, and there is no rigorous research indicating how many biopsies are necessary to snare every actionable mutation.
But a cancer-driving mutation that happens to lie in cells a mere millimeter away from those that were biopsied can be missed. Similarly, cancer cells’ propensity to amass mutations means that metastases, the far-flung descendants of the primary tumor, might be driven by different mutations and therefore need different drugs.
Or, as I like to say: Cancer is complicated. Really complicated. You just won’t believe how vastly, hugely, mind-bogglingly complicated it is. I mean, you may think it was tough to put a man on the moon, but that’s just peanuts to curing cancer, especially metastatic cancer. (Apologies to Douglas Adams.) Because of this, precision medicine as it exists now can lead to what Dr. Don S. Dizon calls a new kind of disappointment when genomic testing fails to identify any driver mutations for which targeted drugs exist because “discovery is an ongoing process and for many, we have not yet discovered the keys that drive all cancers, the therapies to address those mutations, and the tools to predict which treatment will afford the best response and outcome—an outcome our patients (and we) hope will mean a lifetime of living, despite cancer.”
Too true.
None of this is to say that precision medicine can’t be highly effective in cancer. I’ve already described one patient for whom it was. It’s also important to consider that even extra year of life taking a pill with few side effects is “not too shabby,” either, if the alternative is death a year sooner. Prolonging life with good quality is a favorable outcome, even if the patient can’t be saved in the end.
What is precision medicine, anyway?
As I thought about precision medicine during the writing of this post, one thing that stood out to me is that, although precision medicine is rather broadly defined, in the public eye (and, indeed, in the eyes of most physicians and scientists) its definition is much narrower. This narrower definition of precision medicine is the sequencing of patient genomes in order to find genetic changes that can be targeted for treatment, predict the response to therapy of various pharmaceuticals or dietary interventions, or predict disease susceptibility. In other words, it’s all genomics, genomics, genomics, much of it heavily concentrated in oncology. (I concentrated on oncology for this post because it is what I know best.) If you reread the definition from the National Academy of Sciences Committee report, you’ll see that precision medicine is defined much more broadly. Other similar definitions include metabolomics, environmental factors and susceptibilities, immunological factors, our microbiome, and many more, although even a recent editorial in Science Translational Medicine emphasized genomics more than other factors.
In fact, in the most recent JAMA Oncology, there are two articles, a study and a commentary, examining the effect of precision medicine in breast cancer. What is that “precision medicine”? It’s the OncoType DX assay, which is generically referred to as the 21 Gene Recurrence Score Assay.
Basically, this assay is used for estrogen receptor-positive (i.e., hormone-responsive) breast cancer that has not yet spread to the axillary lymph nodes. Twenty-one different genes related to proliferation, invasion, and other functions are measured, and an empirically derived formula is used to calculate a “recurrence score.” Scores below 18 indicate low risk of recurrence as metastatic disease and insensitivity to chemotherapy. Patients with low scores generally receive hormonal therapy but not chemotherapy. Scores over 30 indicate high risk and greater sensitivity to chemotherapy. For such patients, chemotherapy and hormonal therapy are recommended. Patients who score in the “gray” area from 18-30 remain a conundrum, but clinical trials are under way to better define the cutoff point for a chemo/no chemo recommendation. In any case, this study indicates that the use of OncoType DX is associated with decreased use of chemotherapy but because of limitations in the Surveillance, Epidemiology, and End Results (SEER) data set with linked Medicare claims, it wasn’t clear whether this decline was in appropriate patients. In any case, there’s no reason why genomic tests (like the Oncotype DX test) that are rapidly proliferating shouldn’t be considered “precision medicine,” and they are in practice already. Contrary to the image of oncologists wanting to push that poisonous chemotherapy, OncoType DX was designed with the intent of decreasing chemotherapy use in patients who will not benefit. Imagine that.
Conclusion: Medicine that works is just medicine
In the end, I don’t really like the term “precision medicine” that much. It seems to be a term that reminds me, more than anything, of Humpty Dumpty’s famously scornful boast, “When I use a word, it means just what I choose it to mean—neither more nor less.” It’s a sentiment that definitely seems to apply to the term “precision medicine.” To me, when new tests or factors that predict prognosis or response to therapy or suggest which therapies are likely to be most effective are developed and validated, it’s an artificial distinction to link them to genomics, proteomics, or whatever, as well as “big data” and refer to them as “precision medicine.” To me, medicine that works is just “medicine.”